


Multiplex analysis: Up to 5 targets per well.Sensitivity: Detects 1 copy of target sequence in human genomic DNA.Range of excitation/emission wavelengths: 450-730 nm.Optical system: Excitation: 6 filtered LED’s, Detection: 6 filtered photodiodes.Temperature uniformity: ± 0.4 ☌ well-to-well within 10 seconds of arrival at 90 ☌ Temperature accuracy: ± 0.2 ☌ of programmed target at 90 ☌ Key Specifications and FeaturesĬhassis: C1000 Touch / C1000 (older model) Name and Model (Year of Manufacture / Installation)īio-Rad CFX96 Touch Real-Time PCR system (2011/2011), 2 instrumentsīio-Rad CFX96 Real-Time PCR system (2008/2008), 1 instrument General InformationĬFX96 real-time PCR ( PCR =polymerase chain reaction) detection system. Two Touch PCR systems (on the left) and an older PCR system on the right. University of Jyväskylä Teacher Training Schoolĭepartment of Biological and Environmental Scienceįinnish institute for Educational ResearchĬentre for Multilingual academic communicationīack to the list CFX Real-time PCR Systemsīio-Rad CFX Real-time PCR systems.
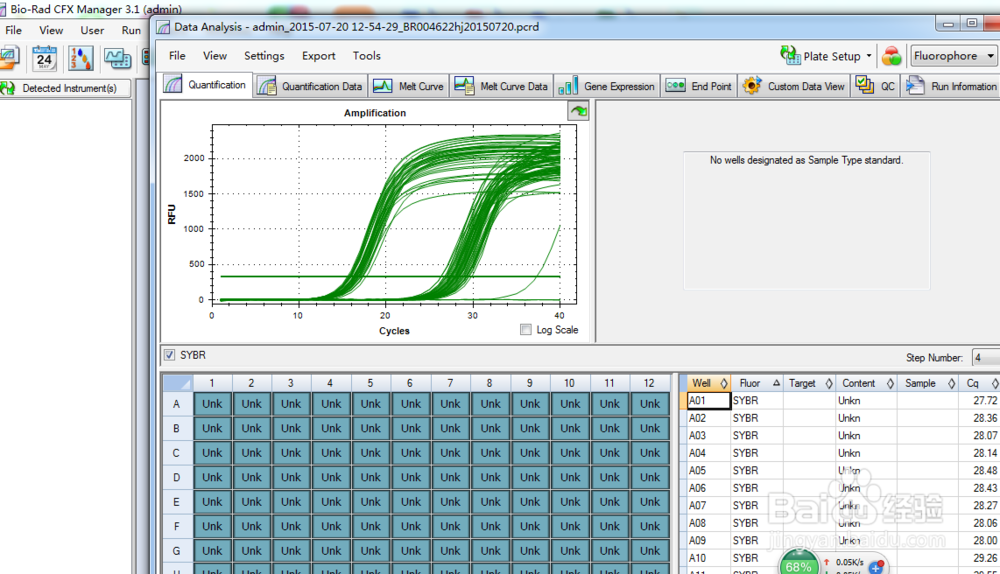
Jyväskylä University School of Business and Economics Matrix normalized worksheet reports normalized signal (against housekeeping genes).Department of Language and Communication Studiesĭepartment of Music, Art and Culture Studiesĭepartment of Social Sciences and Philosophy The CFX manager software automatically performs all deltadeltaCt based fold-change calculations from the uploaded raw threshold cycle data.įold Change worksheet reports test/control (CS-MSCs/BM-MSCs) ratios.Ĭomparative analysis of human bone marrow-derived MSCs and corneal stromal cells Target gene signals normalized to housekeeping genes 2^-deltaCt, where deltaCt = (Ct_Target − Ct_HKG)] The normalization and all the data analysis were performed according to the manufacturers instructions using the CFX Manager software Gene Study ( )įor the normalization it uses the average of three housekeeping genes: TBP, HPRT1 and GAPDH. Quantitative real-time PCR were performed (CFX96 Real-Time System – C1000 Thermal Cycler, Bio-Rad) with 40 cycles at 95☌ for 5 seconds and 60☌ for 30 seconds. From the purified RNA, 10ng was used per sample to synthesize cDNA using the iScript Advanced cDNA synthesis kit (Bio-Rad Laboratories, Temse, Belgium) following the protocol supplied by the manufacturer. QRT-PCR assays were prepared in triplicate in 96-well PrimePCR plates for human MSCs (Mesenchymal stem cells, SAB Target List, Bio-Rad Laboratories, Temse, Belgium). Total RNA was extracted with RNeasy Mini Kit followed by DNase I treatment. All cells were used in the experiments before the 6th passage. When 80% to 90% confluence was reached, cells were passaged using TrypLE (Life Technologies, Gent, Belgium) for 5 minutes. BM-MSCs from a single donor were expanded in DMEM + 10% FBS. GEO help: Mouse over screen elements for information.Ĭell type: corneal stromal mesenchymal stem cellsĬS-MSCs from six donors were seeded after isolation in both Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), as the standard culture condition, and in DMEM supplemented with 10% Human Platelet Lysate (HPL), following our optimized xeno-free culture protocol reported previously (Matthyssen et al.


 0 kommentar(er)
0 kommentar(er)
